How to use AbRSA web service
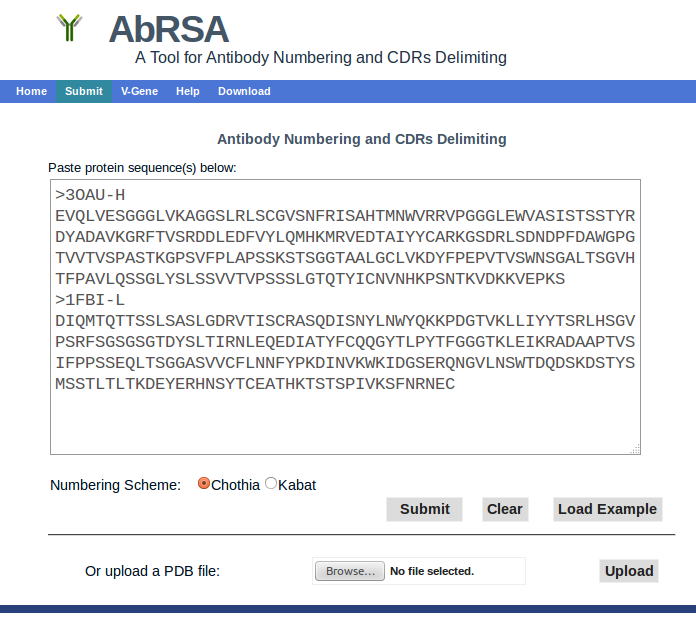
- Please paste query antibody protein sequences, either heavy or light chains, into the textbox. AbRSA supports batch computing by pasting sequences in FASTA format. Examples can be loaded by clicking the "Load Example".
- AbRSA can also number an antibody structure by uploading a PDB format file.
- Please choose the numbering scheme. More information about Kabat and Chothia numbering schemes can be found at http://bioinf.org.uk/abs/
- Please submit the query sequences by clicking button "Submit". Usually the result page will be shown immediately. If you submit hundreds of query sequences, it will take more time to get the results.
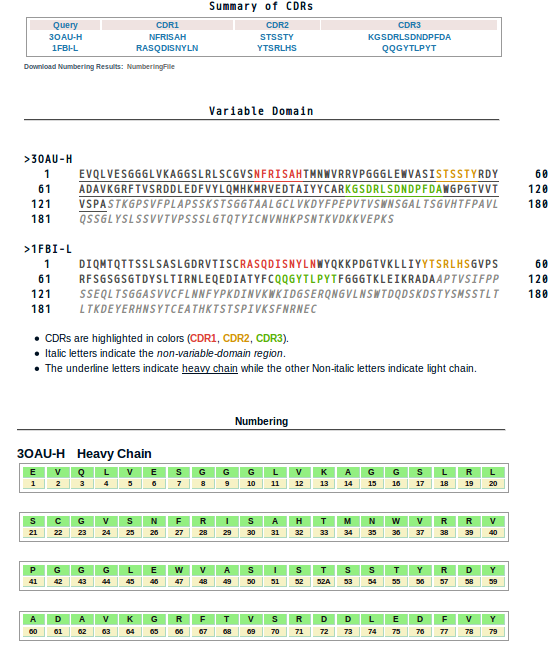
- The results are illustrated in a table (Summary of CDRs) indicating CDR1/2/3 segments of the query sequences. And their positions in sequences are highlighted in red, yellow and green in the following section. Segments which do not belong to variable domains are shown in gray.
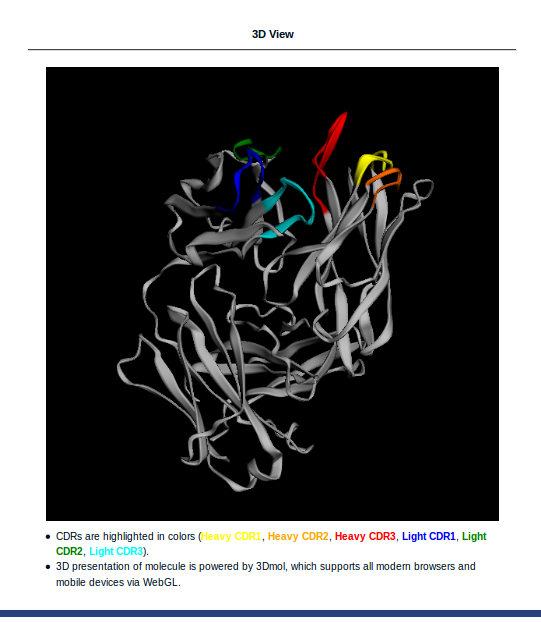
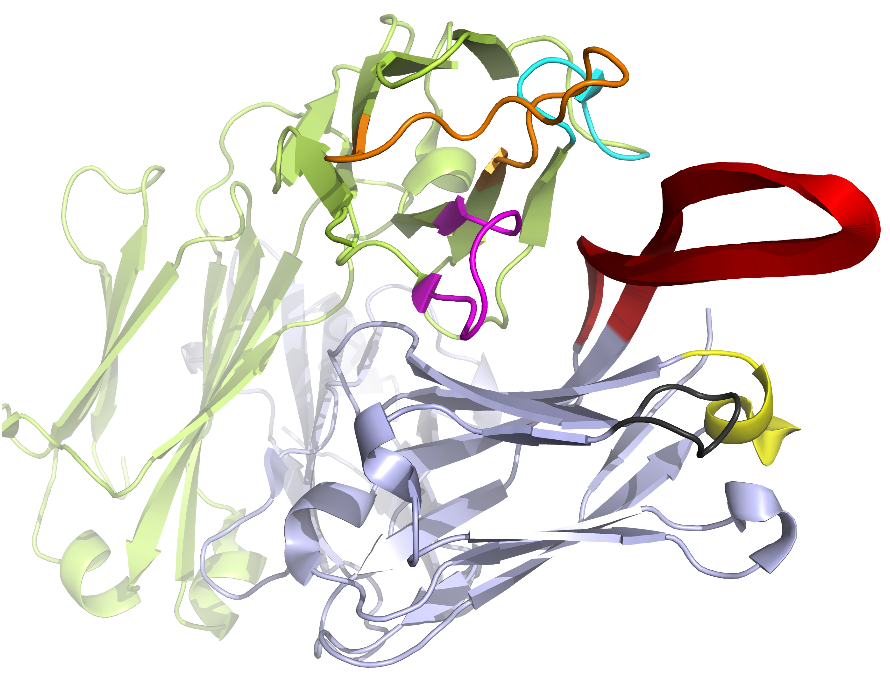
- If an antibody PDB file is loaded, an interactive 3D view will be shown in the 3D View section. CDR1/2/3 segments are highlighted in colors. The 3D view could be rotated, translated, and resized by dragging, scrolling your mouse. 3D presentation of molecule is powered by 3Dmol, which supports most modern browsers and mobile devices via WebGL.
- AbRSA will output warning if the query sequences have low similarities to the antibody consensus sequences.
- If you have more questions, please contact cy_scu@yeah.net.